By Stefano Mammola & Facundo X. Palacio | November 7, 2024
Trait-based ecology is trending! Over the past two decades, the number of trait-based studies has skyrocketed (Figure 1). This pattern suggests that ecologists are increasingly examining species and communities through the lens of functional traits — characteristics such as body size, coloration, dispersal ability, and a species’ role in the food web, all of which are linked to survival and reproduction. Crucially, the use of traits instead of taxonomic information — such as considering butterflies as pollinators with specific morphologies and particular spectra of host plants rather than focusing on the identity of specific butterflies — allows us to infer the processes underlying biological communities. For example, this approach helps us understand how species interact with one another, how biological communities assemble and evolve, and ultimately what ecosystem functions are provided by a given assemblage of species. In the butterfly example above, using traits could allow researchers to infer whether coexisting species differ in size or fly at different times of the day to differentiate their diets and avoid competition, or to map some of the animal-plant feeding associations occurring in a given grass field. On paper, this makes trait-based approaches a holy grail for achieving a mechanistic understanding of ecosystem functioning. But things are not so simple.
 Figure 1. The recent steep increase in the number of published papers using the term ‘functional diversity’ (the diversity of traits in a given species assemblage) compared to ‘phylogenetic diversity’ (the diversity of evolutionary lineages in a given species assemblage). Image credit: Palacio et al. 2022, Ecography
Figure 1. The recent steep increase in the number of published papers using the term ‘functional diversity’ (the diversity of traits in a given species assemblage) compared to ‘phylogenetic diversity’ (the diversity of evolutionary lineages in a given species assemblage). Image credit: Palacio et al. 2022, Ecography
Like any emerging discipline, progress in trait-based ecology has been uneven, unstandardized, and sometimes incoherent. There are now countless methods and metrics for estimating functional diversity based on species traits, often yielding either overlapping or conflicting results, with no clear guidance on which one to adopt to answer a given question. The parallel and often chaotic development of jargon further complicates matters, making it unclear how to label different concepts in trait-based ecology and what processes are genuinely captured by these terms. Questions such as “When can a trait be defined as functional?” and “What is truly captured by different metrics of functional diversity?” are sparking heated debates among experts in the field. Additionally, the data and code supporting studies in trait-based ecology are not always accessible or readily reusable, causing the field to lag behind in terms of reproducibility standards. This begs the question: Moving forward, how can we make trait-based ecology more standardized and reproducible?
In a 2022 paper published in the journal Ecography, we proposed a simple answer to this question. We suggested that researchers working with functional traits could follow a straightforward, general roadmap for conceptualizing and reporting all the typical steps of a trait-based study. This roadmap, which we devised as an 8-steps protocol (Figure 2), is designed to guide researchers in their efforts, from thinking about a relevant research question and study design (steps 1–2) through the collection of data (steps 3–4) and data analysis (steps 5–7), and finally to sharing the data and code supporting the entire procedure using best practices in terms of FAIRness (Findable, Accessible, Interoperable, and Reusable data principles) and reproducibility.
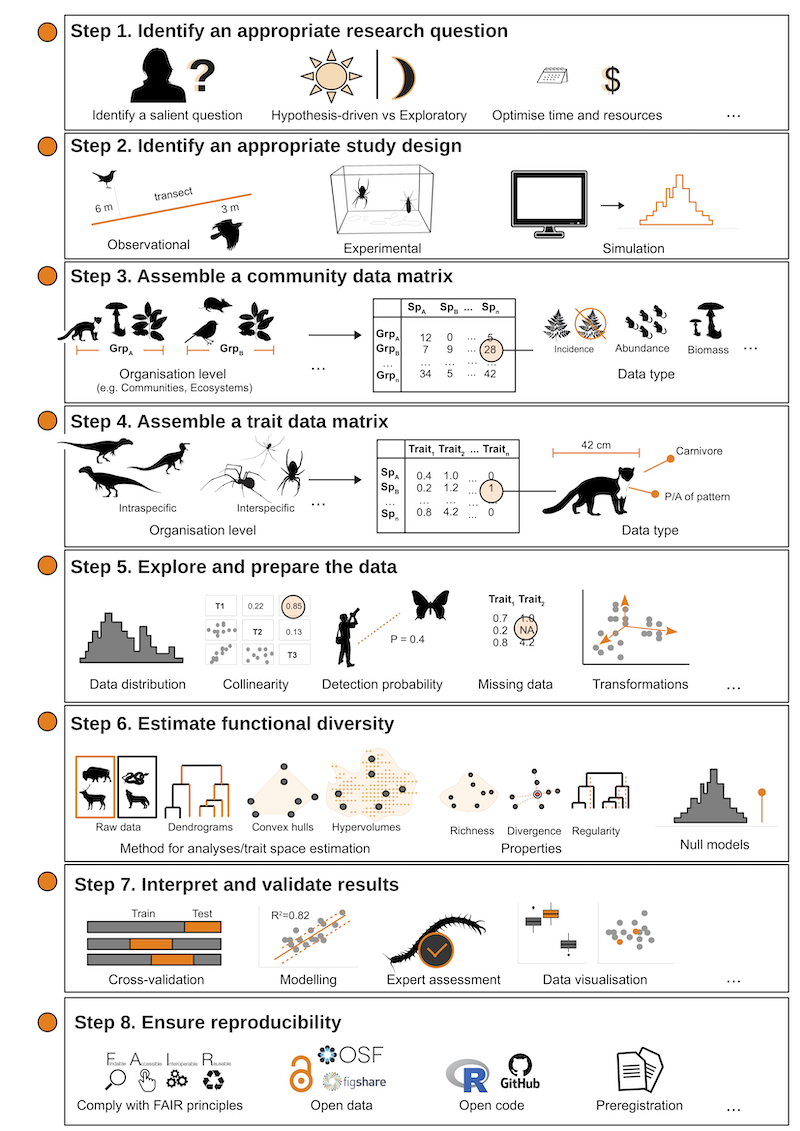 Figure 2. The protocol for reproducible trait-based analyses we proposed. Image credit: Palacio et al. 2022, Ecography
Figure 2. The protocol for reproducible trait-based analyses we proposed. Image credit: Palacio et al. 2022, Ecography
Due to its simplicity and flexibility, we hope for (and encourage!) broad adoption of this protocol. A summary of all eight steps of a given study could be published alongside each paper, for example, as an appendix or supplementary material. We also developed a simple, interactive online template to facilitate the creation of such an appendix, which is available here: https://facuxpalacio.shinyapps.io/stepFD/.
We believe that adopting this simple practice would yield multiple benefits. Firstly, it would help researchers achieve more clarity in their writing of trait-based papers — potentially even helping to alleviate some widely-lamented cases of “writer’s block,” as it is often easier to break down a challenging writing task into smaller steps. Secondly, adopting this protocol would aid readers in navigating complicated scientific papers. Finally, as more and more papers accumulate that rely on this protocol, we envision a future where the extraction of standardized data from multiple papers would be much easier, with obvious benefits such as the possibility of developing synthesis analyses of results from multiple studies (i.e., meta-analyses). So, what are you waiting for? Why not try it out in your next research?
Biography
Stefano Mammola is an ecologist and conservation biologist based at the Molecular Ecology Group (MEG), Water Research Institute of the National Research Council in Verbania, Italy. He specializes in applying various quantitative methods — including trait-based approaches — to understand the dynamics of communities and ecosystems. His primary model systems are cave-dwelling spiders and other mysterious organisms that inhabit the dark depths of subterranean ecosystems.
Facundo X. Palacio is an ecologist based at the Museo de La Plata, Universidad Nacional de La Plata, Argentina. His main interests focus on the evolutionary ecology of plant-bird interactions and the effects of anthropogenic change on bird ecosystem functioning.

